
What is it?
EosFP is a green-to-red irreversibly photoconvertible fluorescent protein. The color change occurs once excited by (near-UV) 390-405 nm light, with the maximal efficiency wavelengths at 390 nm. Prior to excitation the protein has an emission peak of 516 nm. Following excitation, the protein’s emission peak is 581 nm.
Variants of EosFP:
How is it fluorescent?
Photoconvertible fluorescent proteins (PAFPs) contain chromophores that are a formed from a tripeptide within their peptide chains (His65-Tyr66-Gly67). When exposed to UV light, two things happen: (1) Cleavage in the backbone between an amide nitrogen and the alpha carbon of His65. This alpha carbon then forms a double bond with the beta carbon of His65. (2) Extension of conjugated double bonds within the chromophore results in a shift in the fluorescence color spectrum from green to red.
EosFP was discovered by Wiedenmann and colleagues in 2004. A cDNA library was created from L. hemprichii (a coral) and screened for fluorescent clones. In doing so, the researchers discovered a color change. In search of the source of this color change, Wiedenmann and colleagues took coding cDNA fragments and subcloned them into pQE32, and created site-directed mutations before sequencing. All proteins were then expressed in purified to find the protein of interest. The protein which they found to be the cause of the color change was name Eos, after the goddess of dawn in greek mythology.

Although wild-type EosFP is a tetramer, other varients of the protein have been constructed for use in research. The monomer form of the protein mEosFP was constructed through point mutations V123T and T158H. These two mutations interrupted the protein-protein interface of the tetrameric species. However, the protein still matured at a temperature not ideal for research in humans (30 C). Three additional mutations were constructed in the protein (N11K, E70K, and H74N) that allowed for maturity at 37 C. Each of these three mutations improved the protein’s secondary structure. Additionally another mutation was created at His121. With these 4 mutations, mEos2 was introduced.
Computer generated approximation of Eos FP tertiary structure.
Computer generated approximation of the CaMPARI synthetic protein
Since its discovery, EosFP and its variants have been used to optically label single cells (tetrameric form) and to kinetically track single molecules as fused proteins. However, with continued research problems of oligomerization occurred with mEos2 when in high enough concentrations in the cell. This limits its use as a fusion protein in living organisms. Recently, other variants have been made in attempt to solve this problem (mEos3.1 and mEos3.2). Additionally two EosFP’s were combined with calmodulin to create CaMPARI, a protein that has multiple applications in research.
The improvements with mEosFP variants have created a number of improvements in the protein, including higher efficiency at higher temps, a higher emission amplitude, photoconversion at a faster rate and good photostability. Additional properties of EosFP and its variants can be found in the “Things to Know for Research” section.
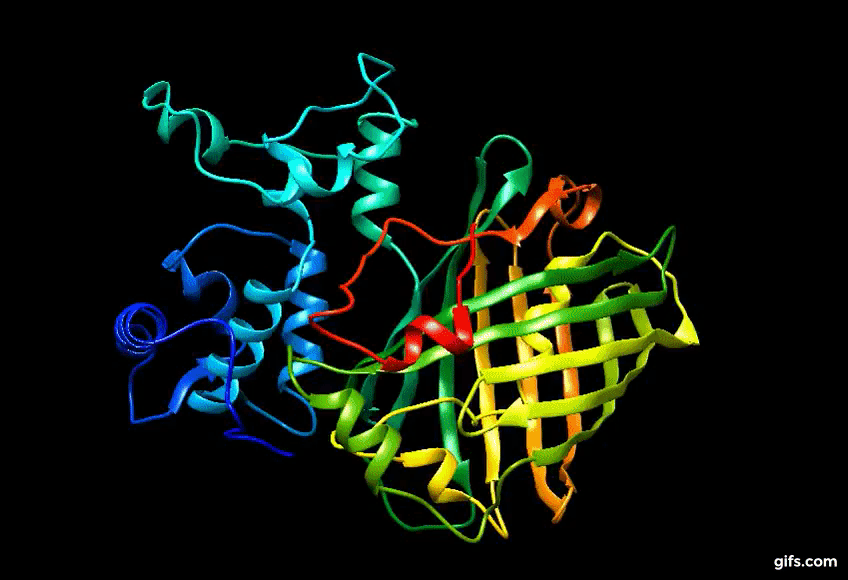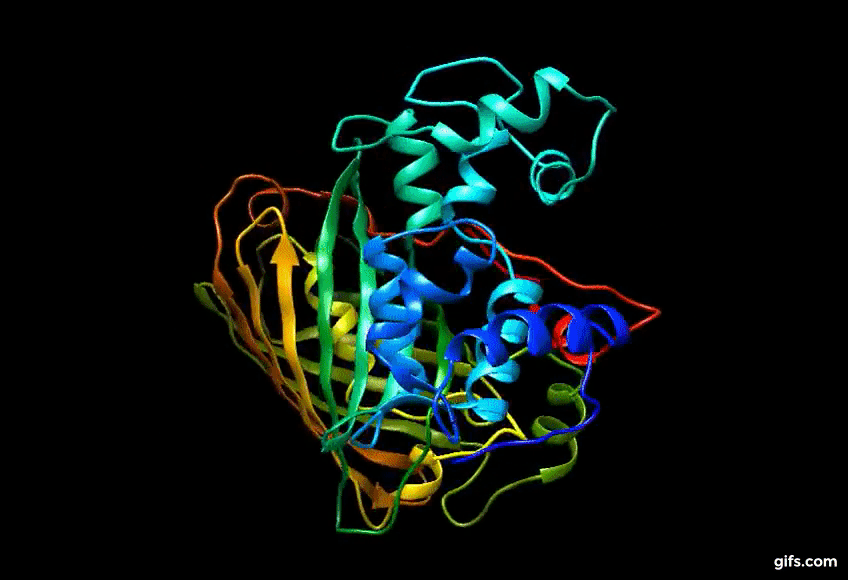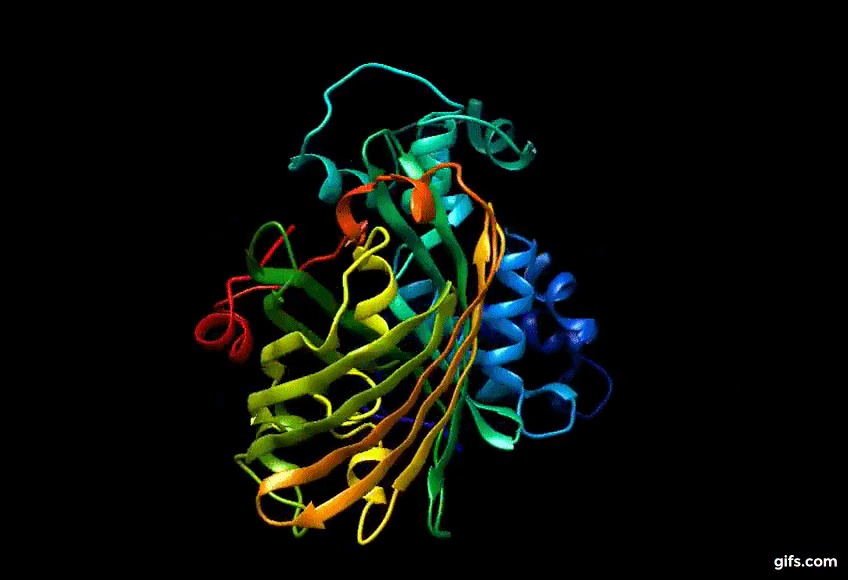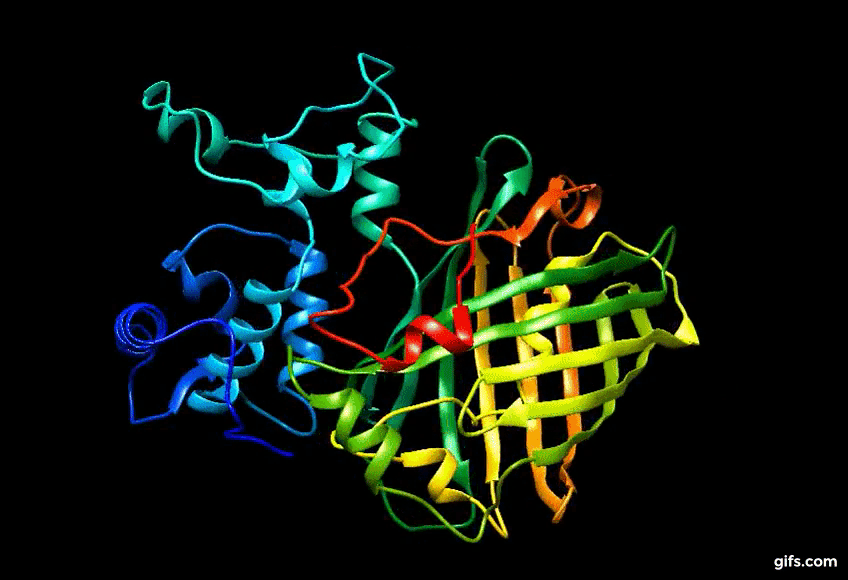
Discovery of Eos FP:
Researchers continued to test the properties of EosFP. The protein was found to be 226 amino acids with a molecular mass of 25.8 kDa and a pI of 6.9. Sequencing analysis revealed a sequence that mostly closely aligned to Kaede (another fluorescent protein), with 84% identical residues. Later studies tested whether the protein converted by backbone cleavage, as Kaede does. Scientists subjected the pre-excited protein, along with the post-excited protein to SDS-PAGE and transferred the gel to a nitrocellulose membrane for a semi-dry blotting procedure. These results showed that backbone cleavage did occur upon excitation of the protein.
To test the expression of EosFP, Wiedenmann and colleagues sub-cloned the cDNA of the protein and its variants into expression vector pcDNA3 with and without Notch protein (mNotch-1-IC) and recognition signal binding protein 2N (RBPO-2N). Additionally some vectors were also incorporated with the cDNA for mitochondrial targeting signal from subunit VIII of cytochrome c oxidase. Fusions proteins were created from these vectors and compared to fusion proteins made with GFP (EGFP), as well.




